[Work Log] (Two day) Markov Sampling (ctd). Implementing, testing, optimizing
Note: changes to params.
Resuming from yesterday's discussion about Markov sampling. I was concerned about piecewise sampling of interpolated values with insufficient data being used. After some thought, realized there's a better approach: construct blocks from input indices, not output indices.
Secondly: only use the markov approach when there's too much data to eat at once. Those cases are also the ones with the least probability of poor-data issues.
Re-implemented (curve_tree_ml_5.ml), geting weird results. Huge negative eigenvalues
symptom: 30 output indices, 1000 output indices symptom: K has 26k elements!
This code is a mess, proving hard to debug. I'm going to roll back to version 4, use ideas developed in v5 to implement Markov sampling.
Done. No errors, but results aren't great. Is it possible the markov blanket is wrong, or maybe we're misusing previous data?
...
I think I found it... wasn't conditioning on previous sampled values.
Seems to be fixed now. Next on to timing and tuning
Profiling / Optimizing
- attachment covariance
- markov order
- block size
Constructing Attachment covariance
construct_attachment_covariance_3 is still a bottleneck. Need to investigate some of the suggestions from a few days ago.
Only computing Cov_star_star once (exploiting stationarity in temporal GP) helps somewhat.
Grouping "sibling" object construction should help a lot too.
Markov order
Markov order as low as 10 doesn't seem to negatively affect results.
Need to crop observations before the earliest sampled point we're conditioning on.
Timing
Before: 17.0s (total, 7.5s on inversion)
After: 16.5 (7.2s)
Tiny improvement, but at no cost so who's complaining?
Block Size
Not much to say here... 100 seems to be optimal.
Markov Order
Plotted maximum posterior for various markov orders to compare reconstruction quality.
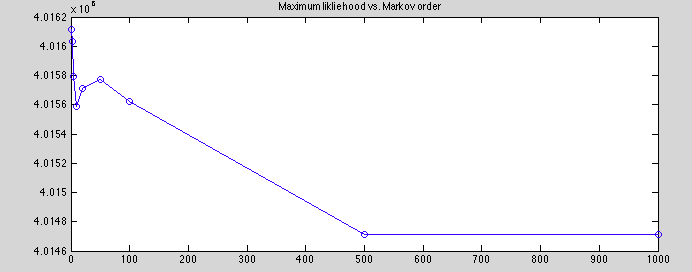
range: 1.40e3
(unweighted) mean: 4.0156
std deviation: 510.0
the mean should be taken with a grain of salt, Some chaos below 500, but the scale of the fluctuations is actually relatively small, 0.035%. We should note that in MCMC, its the absolute fluctuations that are significant, so percent error can be deceptive. But even so, I think these results are pretty good.
It's interesting that markov order of 10 is only slightly better than markov order of zero! Also surprising that between 10 and 500, error increases.
Should compare variance here vs. variance of log-likelihood w.r.t. posterior.
Result of 20 posterior samples (low markov order)
markov order = 10
block size = 100
ll std deviation = 2.33e3
ll mean: 4.0153e6
Result of 20 posterior samples (high markov order)
markov order = 500
block size = 100
ll std deviation = 2.17e3
ll mean: 4.0132e6
The log-likelihood standard deviation are high relative to the error range in the graph above. This is weak evidence that the variance in the graph above is dues to small-scale instabilities in the pixel likelihood. That is to say, tiny insignificant changes to the 3D model can cause noticible fluctuations to the pixel likelihod, due to it's nonlinearities. To some extent, what we really want is the expected value of the likelihood over the entire set insignificant 3D perturbations. So if the deviations we see due to markov-order are simply observations of this phenomenon, they can be safely ignored, because they are in any scenario.
Let's test by re-running the low-markov order test with lots of samples (200 instead of 20) and see if the ll mean approaches that of the high-markov-order test. This is also a decent stress test for the likelihood server.
Result of 200 posterior samples (low markov order)
markov order = 10
block size = 100
ll std deviation = 2.5603e3
ll mean: 4.0133e6
Yep, we're getting closer to the high markov order results for ll mean. I think we can drop this issue for now and be okay with a markov order of 10-20. It is still surprising that markov order of zero seems to work so well. I guess future data doesn't add too much if the present data is sufficient.
Bottlenecks
Inversion: 7.1s buildling covariance: 2.5s one_d_to_three_d: 1.5s blkdiag: 1.2s other: ~3.5s
Inversion
Only need to invert once for all views... Actually, not true because inversion contains view-specific sample data. But a large block of the matrix is unchanged between views. Implementing optimization...
...
Down to 14.7 seconds.
...
Using sparsity and suppressing output, down to 11.5s. (8s when not in profile mode)
However, getting some numerical issues (small-magnitude negative eigenvlaues); probably will do better if we use a symmetric equation and invert using cholesky.
construct_attachment_covariance is now the bottlneck, taking a full 35% of run time.
Some options:
- cache intermediate values here and return them as described in the previous post.
- group together calls to "build_sibling_object"
- avoid computing self-covariance when its available in prior_K
- implement one-pass version for symmetric matrices
- precompute self-covariance for all views in one call.
Implemented 2, 3, and 4, reduced running time to 8.7s.
Implemented 1, reduced to 8.4s.
Implemented 5, now construct_attachment_covariance is not a bottleneck. 8.3s.
Not many strong bottlenecks now. Some opportunities to avoid build_cov_ and one_d_to_three_d like we did in curve_tree_ml_5_bak. Also, using a symmetric formula for posterior might help here.
Tweaked build_cov_ to skip an inner loop if it would be a no-op. Down to 7.8s.
Set block size to 100 (had forgotten to change back yesterday, was 120) 7.6s.
Rearranged matrix multiplication. 7.4s
Mex'd one_d_to_three_d utility function. 6.9s
Problem. Samples look bad. Overall shape is retained, but rough, with discontinuities.
Switched off block matrix inversion, results look good, but back to 8.3s
Found bug. Indexing problem was causing previous samples to be ignored. Fixed, and its better than before, but still getting some non-negligible eigenvalues and occasional discontinuities in samples.
Summary
Originally took 275s, now 8.3s in profile mode, 5.x s in regular mode.
TODO (new)
- running time vs. number of samples to determine affect of likelihood server bottleneck (use GPU likelihood for real-world estimate)
- profile/optimize likelihood server directly?
- Symmetrize formula and use cholesky for inversion.
